Dissecting Our Cellular Power Plants
A new study sheds light on how mitochondrial proteins are synthesized, providing new targets to fight childhood neurological diseases
By Josh Baxt
Photography by Tom Salyer

M
itochondria make multicellular life possible. These tiny organelles convert glucose into energy, making them critically important for cell survival. Dysfunctional mitochondria can cause devastating diseases, frequently with childhood onset, but until recently they have been difficult to study.
Now, in a paper published as the cover story in the February 19 issue of the journal Science, an international team, including researchers from the Miller School of Medicine and colleagues Alexey Amunts, Ph.D., associate professor of biochemistry and biophysics at Stockholm University in Sweden, and Brendan Battersby, Ph.D., a principal investigator in the Institute of Biotechnology at the University of Helsinki in Finland, has delineated several structures associated with mitochondrial protein synthesis. These findings could help scientists understand the mechanisms that go wrong in some of the deadliest mitochondrial diseases, as well as offer potential therapeutic targets.
“Many of these diseases are embryonically lethal,” said Antonio Barrientos Ph.D., professor in the departments of Neurology and Biochemistry and Molecular Biology and co-lead author on the study. “Even if the kids are born, they often die after only a few years. Now, we have uncovered a basic mechanism that drives pediatric mitochondrial disease.”
An ancient story
This story begins about a billion years ago, when single cell organisms may have partnered with symbiotic bacteria able to use oxygen for energy production. It was a good deal for both organisms — the bacteria got food, and the cells received energy. However, as a result of this ancient pairing, mitochondria have their own DNA, separate from the genome in the cellular nucleus.
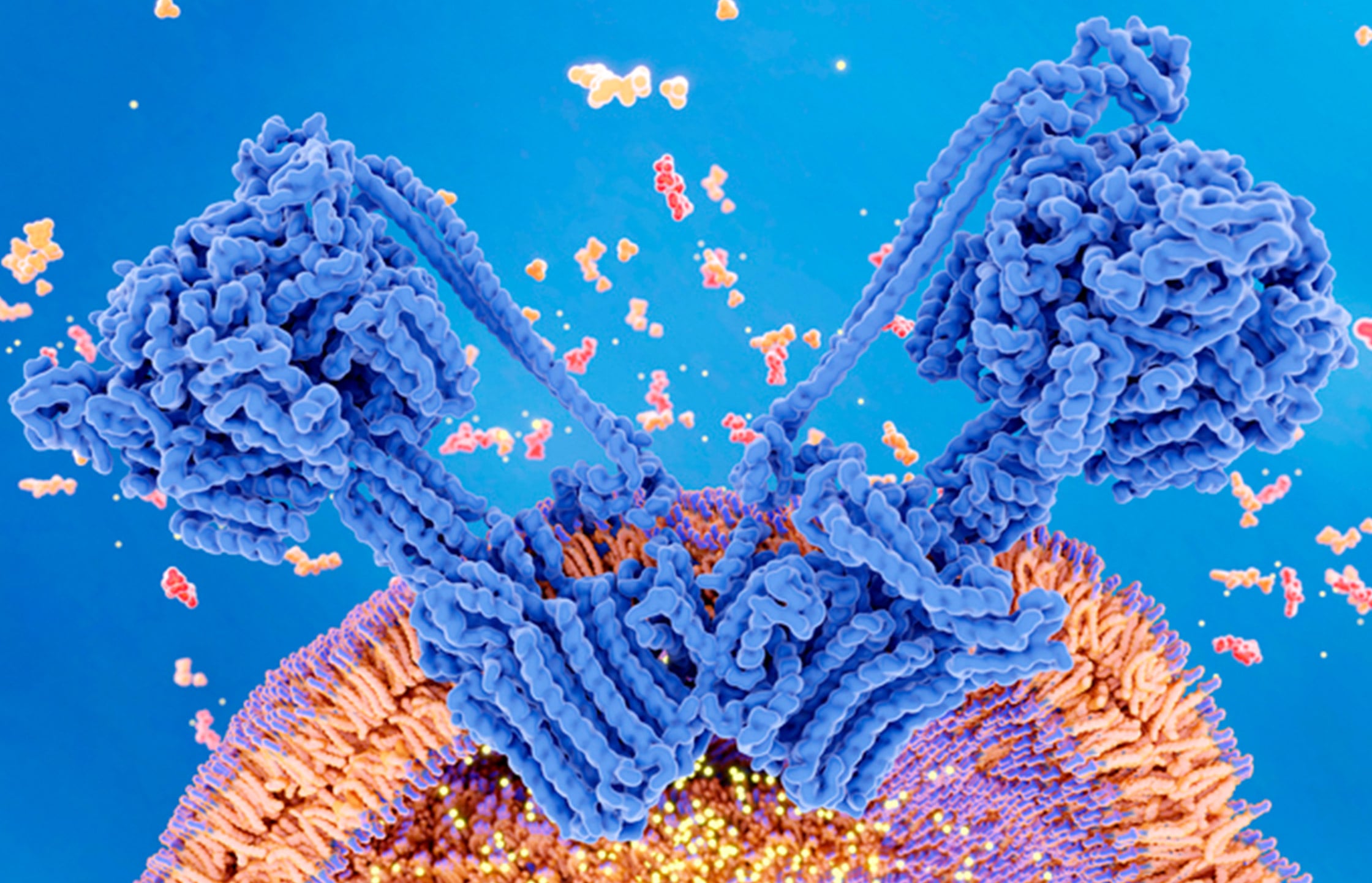
Like nuclear DNA, mitochondrial genetic material is transcribed into RNA, which is translated in mitoribosomes to produce 13 mitochondrial proteins. The Barrientos lab has been working for a decade to understand how these mitochondrial ribosomes are assembled and function. For this specific project, he worked with Austin Choi, a graduate student in neuroscience and co-first author of the manuscript, and Priyanka Maiti, Ph.D., a postdoctoral fellow.
This is an essential process, but until now, no one had fully observed how these mitoribosomes are structured to facilitate the insertion and distribution of their finished proteins in the mitochondrial membrane. The new paper sheds light on these processes.
Austin Choi

The study revolves around a protein called OXAIL-1, which binds to the protein-making machinery and shepherds completed proteins into the mitochondrial membrane. This is a critical step. Because these proteins are hydrophobic, they can only be effective in the lipid (fatty) membrane. If OXAIL-1 doesn’t do its job, mitochondrial proteins remain in water, lose their shape and become useless.
A good fit

“This fits in with what we already knew,” Dr. Barrientos said. “Mutations in OXAIL-1 cause cardio- and encephalomyopathies, which are devastating diseases. These proteins cannot be inserted into the membrane properly, creating a big mess. This gives us tremendous insights into the pathogenic mechanisms for these mitochondrial diseases.”
Prior to this study, no one had been able to image how mitochondrial ribosomes bind to the mitochondrial membrane and insert completed proteins into that membrane. Using cryo-electronic microscopy, the team diagrammed this process in detail, giving researchers new information that could eventually lead to better treatments for mitochondrial diseases.
“Now that we know how this system works a little better, we can identify some targets for therapeutic intervention,” Dr. Barrientos said. “Targeting mitochondrial translation is emerging as a novel therapeutic tool to combat mitochondrial encephalomyopathies and metabolic disorders, an avenue we will continue investigating.”![]()
Priyanka Maiti, Ph.D.